Scientists reveal the mechanism of rifampin inactivation by rifampin phosphotransferase
Rifampin (RIF), the best-known member of rifamycins, is a first-line drug for the treatment of tuberculosis, and also used to treat other bacterial infections. Extensive use of rifamycins has led to the development of bacterial resistances, which is a great threat to the prevention and treatment of infectious disease. Understanding the molecular mechanisms is essential to overcome these resistances. RIF phosphotransferase (RPH) is an antibiotic-resistance protein family and was identified to inactivate RIF and other members of rifamycins by phosphorylation at the cost of ATP. However, the underlying mechanism remains unknown.
In a recent study, researchers from Dr. ZHANG Peng’s group solved the structures of RPH from the pathogen Listeria monocytogenes (LmRPH) in different conformations. LmRPH comprises three domains: an ATP-binding domain (AD), a RIF-binding domain (RD), and a catalytic His-containing domain (HD). Structural analyses reveal that the ATP-binding site in AD and the RIF-binding site in RD are far from each other, and the C-terminal HD can swing between the AD and RD, like a toggle switch, to transfer phosphate. In addition to its catalytic role, the HD can bind to the AD and induce conformational changes that stabilize ATP binding, and the binding of the HD to the RD is required for the formation of the RIF-binding pocket. A line of hydrophobic residues forms the RIF-binding pocket and interacts with the 1-amino, 2-naphthol, 4-sulfonic acid and naphthol moieties of RIF. The R group of RIF points toward the outside of the pocket, explaining the low substrate selectivity of RPH. Four residues near the C21 hydroxyl of RIF, His825, Arg666, Lys670, and Gln337, were found to play essential roles in the phosphorylation of RIF; among these the His825 residue is phosphorylated in the presence of ATP and may function as the phosphate acceptor and donor. Base on the results above, researchers revealed the molecular mechanism of RIF phosphorylation catalyzed by RPH and the phosphorylation may make rifamycins unable to fit into its binding pocket on the target (bacteria RNA polymerase), losing their antibacterial activity. This study will guide the development of a new generation of rifamycins.
This study entitled “Structural basis of rifampin inactivation by rifampin phosphotransferase” was published online in PNAS on March 21, 2016 (http://www.pnas.org/content/early/2016/03/18/1523614113.full). This work was funded by the National Natural Science Foundation of China, Ministry of Science and Technology of China and Chinese Academy of Sciences.
AUTHOR CONTACT:
Zhang Peng
National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, CAS.
Tel: 86-21-54924219;
Email: pengzhang01@sibs.ac.cn
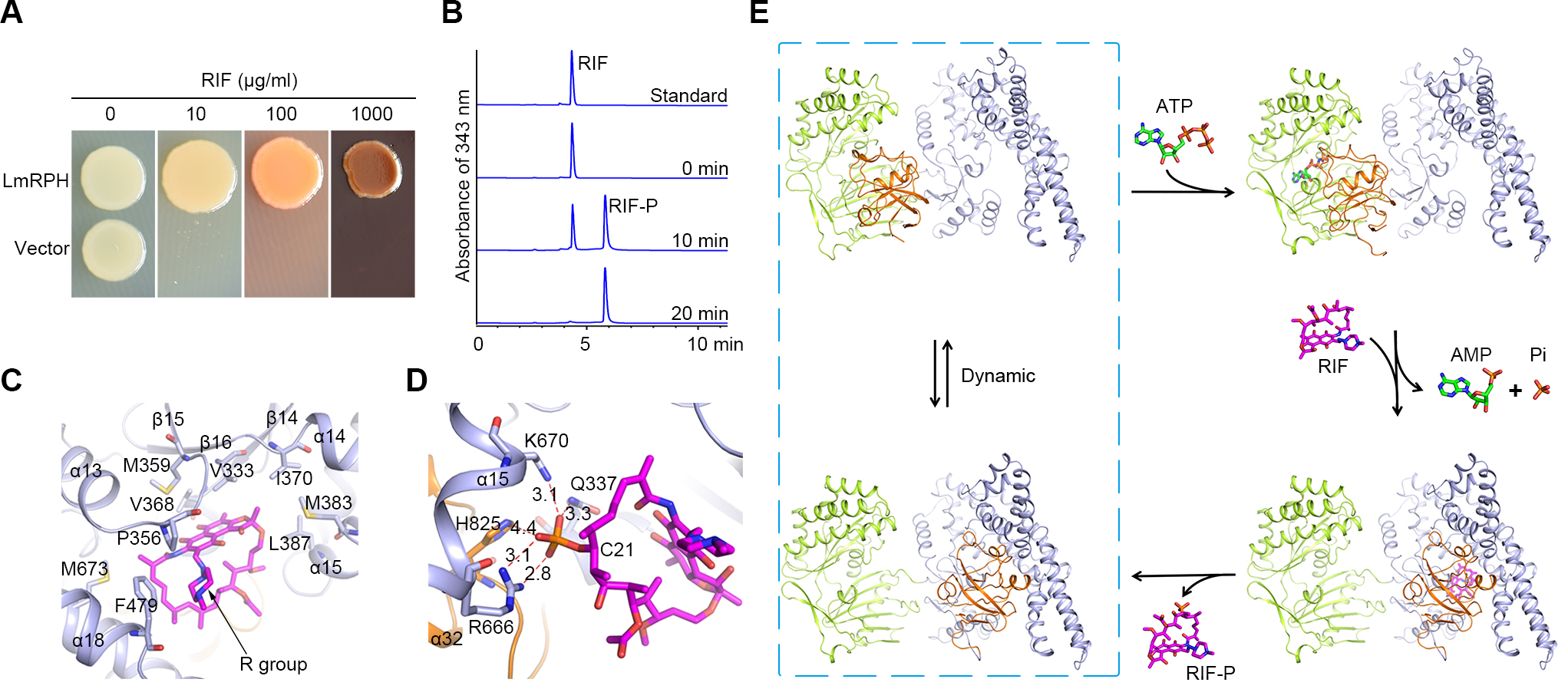
A, LmRPH leads to RIF resistance assayed using E. coli; B, In-vitro enzyme activity of LmRPH detected by HPLC; C, RIF-binding pocket (phosphorylated RIF is shown in magenta stick model) ; D, Catalytic site of LmRPH; E, Catalytic mechanism of LmRPH mediated rifamycins phosphorylation.
