CIBs Form Heterodimers to regulate the CRY2-dependent Flowering
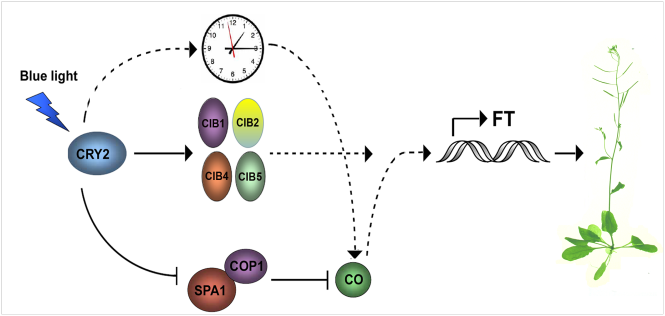
The timing of the flowering is critical for reproductive success for the plants. Arabidopsis thaliana blue light receptor cryptochromes (CRYs) mediate light control of flowering time by interacting with CIB1 (CRY2-interacting bHLH1) in response to blue light. However, it remains unclear how the blue light dependent CRY2-CIB1 interaction affects the FT transcription.
A group led by Dr Liu Hongtao from the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences provides evidence that CIBs function redundantly in regulating CRY2-dependent flowering, and that different CIBs form heterodimers to interact with the non-canonical E-box DNA in vivo.
They showed that CRY2 also interacts with at least CIB5, in response to blue light, but not in darkness or in response to other wavelengths of light. Their genetic analysis demonstrated that CIB1, CIB2, CIB4, and CIB5 act redundantly to activate the transcription of FT and that they are positive regulators of CRY2 mediated flowering. More importantly, CIB1 and other CIBs proteins form heterodimers, and some of the heterodimers have a higher binding affinity than the CIB homodimers to the non-canonical E-box in the in vitro DNA-binding assays. Their result explains why in vitro CIB1 and other CIBs bind to the canonical E-box (G-box) with a higher affinity, whereas they are all associated with the non-canonical E-boxes at the FT promoter in vivo.
Consistent with the hypothesis that different CIB proteins play similar roles in the CRY2-midiated blue light signaling, the expression of CIB proteins is regulated specifically by blue light.
This work entitled “Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in Arabidopsis” has been published online in PLoS Genetics on October 10th. The research was funded by NSFC and CAS.
A working model depicting the function of CRY2 and CIBs in the regulation of flowering time (Image by De Liu Hongtao’s group)